AIMS #CM01: Spontaneous beating wild type hiPSC-ventricular cardiomyocyte field potential assay for compound testing and/or disease modeling
Last revised: Dec 2, 2024

Together with key opinion leaders in academia and in industry, we have incorporated [1] our experiences with hiPSC-cardiomyocytes in our respective labs, [2] our work with international consortia developing hiPSC-cardiomyocyte assays (CiPA and JiCSA), and [3] published data, to develop a proposed standard. This standard focuses on the hiPSC-cardiomyocyte spontaneous beat rate, features of the cardiac waveform (depolarization spike amplitude and field potential duration), and the synchronization of activity in the syncytia. Cell performance data from leading commercial sources of hiPSC-cardiomyocytes with respect to this proposed standard are also presented for reference.
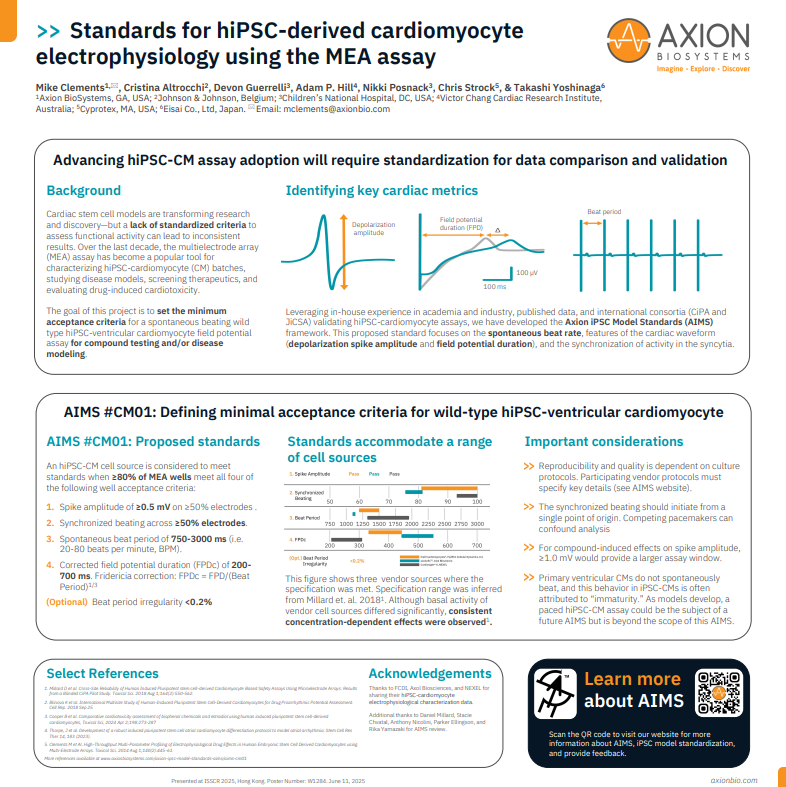
Standards for hiPSC-derived cardiomyocyte electrophysiology using the MEA assay
Learn more about AIMS #CM01 as presented at ISSCR 2025.
>> A wild type hiPSC-ventricular cardiomyocyte batch of cells is considered to have met the Axion Induced Model Standard (AIMS) when:
≥80% of the wells seeded with these hiPSC-ventricular cardiomyocytes meet all four of the following well acceptance criteria:
- For convenience, when using a 24-well plate this would be ≥20 wells, for a 48-well plate ≥39 wells, and a 96-well plate ≥77 wells.
- ≥80% was set as the threshold because lower than this level makes the cost of running the assay prohibitively expensive (cost/well).
- Wells that don’t meet these criteria due to operator error (e.g., due to an error in loading the cells into the MEA plate) will be discarded from this percentage success rate calculation as the impacted well is not an accurate reflection of the cell type’s performance.
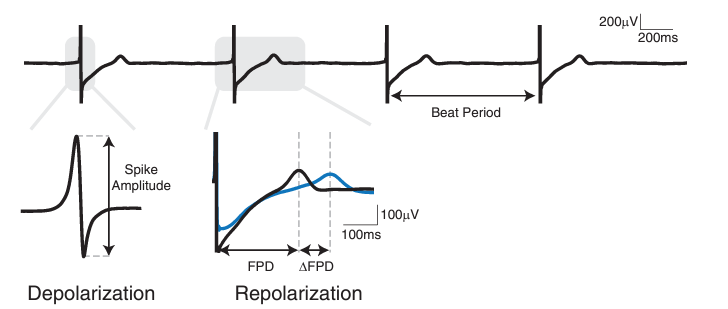
1. Spike Amplitude of ≥0.5 mV on ≥50% electrodes in a well.
- 0.5 mV was set as the threshold for spike amplitude as this allows for reliable capture of the spontaneous hiPSC-CM activity in baseline and could also accommodate a significant compound-induced decrease in spike amplitude.
For studies focused on compound-induced effects on spike amplitude, a spike amplitude of ≥1.0 mV would be preferable to provide a larger assay window.
>> Where to find in AxIS software:
In AxIS Navigator, change the “Detection Threshold” parameter to 500 µV in the Cardiac Beat Detector Settings prior to saving the Advanced Metrics file from the Statistics Compiler. Note: the default Detection Threshold is 300 µV. To determine if a well meets this criterion, look at the “Total Active Electrodes” metric in the Well Averages section of the Advanced Metrics CSV file. A well meets this criterion if the number of active electrodes reported here is greater than or equal to half of the total number of electrodes in the well. For convenience, when using a 24- or 48-well plate, this would be ≥8 electrodes, and when using a 96-well plate, ≥4 electrodes.
2. Synchronized beating across ≥50% electrodes in a well.
- Electrodes in the array are considered to have synchronized beating if within a window of 30 consecutive beats, consistently, the first and last beat across electrodes occur within 30 ms of each other.
- For convenience, when using a 24-well plate this would be ≥8 electrodes, for a 48-well plate ≥8 electrodes, and a 96-well plate ≥4 electrodes.
The synchronized beating should initiate from a single point of origin, where the interelectrode beat time delay increases monotonically as a function of distance from the origin. More than one patch of spontaneous beating iPSC-CMs in a well is undesirable and is indicative of a poorly coupled syncytium. The competing spontaneous activity can confound the analysis of the results. Consequently, if a well has two or more independent patches of spontaneously beating iPSC-CMs in the baseline recording, this well should be discarded.
>> Where to find in AxIS software:
In AxIS Navigator, verify that the “Min Active Electrodes” parameter in the Cardiac Beat Detector Settings is set to 50% prior to saving the Advanced Metrics file from the Statistics Compiler. Note: this is the default setting. To determine if a well meets this criterion, look at the “Propagation Consistency (%)” and Conduction Velocity Mean (mm / ms)” metrics in the Well Averages section of the Advanced Metrics CSV file. A well does not meet the Synchronized Beating criterion if the Propagation Consistency = 0 and no Conduction Velocity is reported.
3. Spontaneous Beat Period of 750-3000 ms (i.e. 20-80 beats per minute, BPM).
- The acceptable beat period range is wide. However, rationale for this range was inferred from the CiPA Pilot Study [Millard et. al. 2018] which used hiPSC-CMs from two vendors with spontaneous beat periods at the two extremes of this range (ref. Figure 2). Although the spontaneous activity of these two cell types differed significantly, consistent concentration-dependent effects were observed for both cell types (ref. Figure 5).
- Working with hiPSC-CMs with spontaneous beat periods greater than 3000 ms can be impractical due to the longer recording periods required to capture sufficient beating events for analysis data sets.
- Primary ventricular CMs should not spontaneously beat, and the spontaneous beating observed in hiPSC-ventricular CMs is often attributed to their “immaturity.” Efforts to enhance the “maturation” of hiPSC-ventricular CMs are ongoing and can result in spontaneous beat periods greater than 3000 ms. For assays involving these hiPSC-CMs, external pacing (via electrical- or light-induced means) will probably be required to make these cells more practical to work with. A paced hiPSC-CM assay could be the subject of a future AIMS but is beyond the scope of this AIMS.
Note: AIMS, #CM01, is specifically for hiPSC-ventricular CMs and not hiPSC-atrial CMs. Studies have shown that hiPSC-atrial CMs can often have a spontaneous beat period less than 750 ms [Thorpe et. al., 2023] (ref. Figure 2).
>> Where to find in AxIS software:
To determine if a well meets this criterion, look at the “Beat Period Mean (s)” metric in the Well Averages section of the Advanced Metrics CSV file from AxIS Navigator. A well meets this criterion if the mean beat period reported here is between 0.75 and 3.0 s.
4. Corrected Field Potential Duration (FPDc) of 200-700 ms.
- Where the Field Potential Duration (FPD) is corrected using the Fridericia correction:
- FPDc = FPD/Beat Period0.33
The acceptable FPDc range is wide. However, rationale for this range was inferred from the CiPA Pilot Study [Millard et. al. 2018] which used hiPSC-CMs from two vendors with FPDc at the two extremes of this range (ref. Figure 2). Although the FPDc of these two cell types differed significantly, consistent concentration-dependent effects were observed for both cell types (ref. Figure 5).
>> Where to find in AxIS software:
This metric can be found in the Well Endpoints CSV file exported from Axion’s Cardiac Analysis Tool. To determine if a well meets this criterion, look at the “FPDc (Fridericia ms)” metric in the Measurement section of the Well Endpoints CSV file from the Cardiac Analysis Tool. A well meets this criterion if the FPDc reported here is between 200 and 700 ms.
>> Additional points to consider:
A. Spontaneous beat regularity
Regularity of spontaneous hiPSC-CM beating was identified as an important factor in the quality of a hiPSC-CM recording. However, this parameter (sometimes measured as coefficient of variance of the beat period) is less commonly reported in the literature [Clements & Thomas, 2014] (ref. Figure 3). Consequently, the Review Team did not feel comfortable setting acceptance criteria relating to beat regularity at this time, but this will be something to consider should this AIMS be refined in the future.
B. Cell culture protocol
The reproducibility and quality of the hiPSC-CM model’s functional activity is dependent on the cell culture conditions. Consequently, the cell provider must specify the following details in their recommended protocol:
- Temperature of the environmental chamber in the Maestro MEA system (e.g., 37.0oC).
- CO2 concentration in the environmental chamber in the Maestro MEA system (e.g., 5.0%).
- Cell culture medium that the MEA recordings should be performed in (e.g., RPMI 1640/B27 medium).
- When the cell culture medium should be refreshed prior to the MEA recording (e.g., a full-medium change 4 hours prior to baseline acquisition).
- Equilibration time given between docking the MEA plate on the Maestro MEA system and the start of the recording (e.g., 10 minutes).
- The experimental window, i.e. the specific timeframe within which the hiPSC-CMs will perform as claimed (e.g., between 7-14 days post-thaw; or day 25-30 post-differentiation; etc.).
- Recommended appropriate positive and negative controls for the response of interest, with expected responses detailed for the former (e.g., application of the hERG K+ channel blocker, E-4031, at 100 nM should result in a ≥20% increase in the FPDc).
>> Below is a list of cell models that have been self-certificated by the vendor to meet the AIMS #CM01 criteria:
iCell Cardiomyocytes2, 01434, FUJIFILM Cellular Dynamics, Inc.
Catalog Number(s): R1017 (5M cells plus media); R1059 (1.25M cells plus media); C1016 (5M cells only); C1058 (1.25M cells only)
AIMS CM#01 requirements:
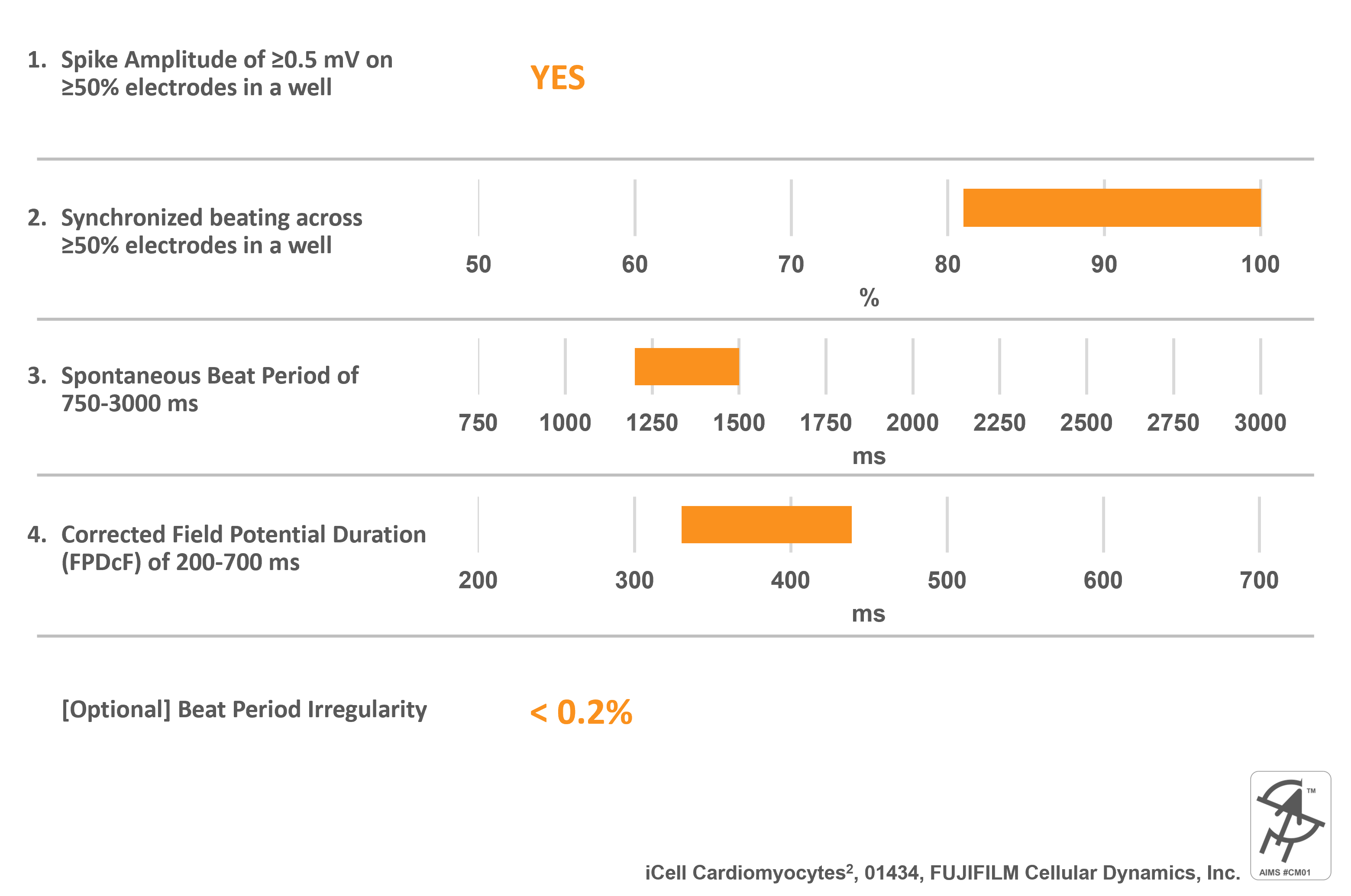
Cell specific information:
- Differentiated from an FCDI reprogrammed human iPSC line
- Apparently Healthy Normal Caucasian Female Donor 01434
- >95% pure Human Cardiomyocytes
- Mixture of Ventricular, Atrial, and Nodal cells
- Preferred hiPSC-CM model used and validated during the CiPA studies
Recommended protocol:
Download Protocol
- Temperature on Maestro Pro MEA system is 37 ºC and CO2 concentration is 5.0%.
- Cells should be maintained in iCell Cardiomyocytes Maintenance Medium (M1003). Cells can be assayed in this same medium, otherwise serum-free options are available including iCell Cardiomyocytes Serum-Free Assay Medium (M1038) or iCell Cardiotox Assay Medium (M1039).
- Recommended assay window for testing iCell Cardiomyocytes2 is Day 4-7 post-thaw.
- On the day of assay, perform 100% media change and wait for 2-4 hours to begin testing.
- Equilibrate MEA plate for 10 minutes prior to baseline recording on MEA system.
- Recommended positive control compounds are E-4031 (100 nM) or Dofetilide (30 nM). hERG block always prolongs FPD by at least 25-40% after 30 min post-dose recording. Negative control is 0.1% DMSO.
Recommend select publications:
- Millard D et. al. Cross-Site Reliability of Human Induced Pluripotent stem cell-derived Cardiomyocyte Based Safety Assays Using Microelectrode Arrays: Results from a Blinded CiPA Pilot Study. Toxicol Sci. 2018 Aug 1;164(2):550-562. doi: 10.1093/toxsci/kfy110.
- Blinova K et. al. International Multisite Study of Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes for Drug Proarrhythmic Potential Assessment. Cell Rep. 2018 Sep 25;24(13):3582-3592. doi: 10.1016/j.celrep.2018.08.079.
- Patel D et. al. Assessment of Proarrhythmic Potential of Drugs in Optogenetically Paced Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Toxicol Sci. 2019 Jul 1;170(1):167-179. doi: 10.1093/toxsci/kfz076.
axoCells™ Human iPSC-Derived Ventricular Cardiomyocytes, Axol Bioscience
Catalog Number(s): ax2508
AIMS CM#01 requirements:
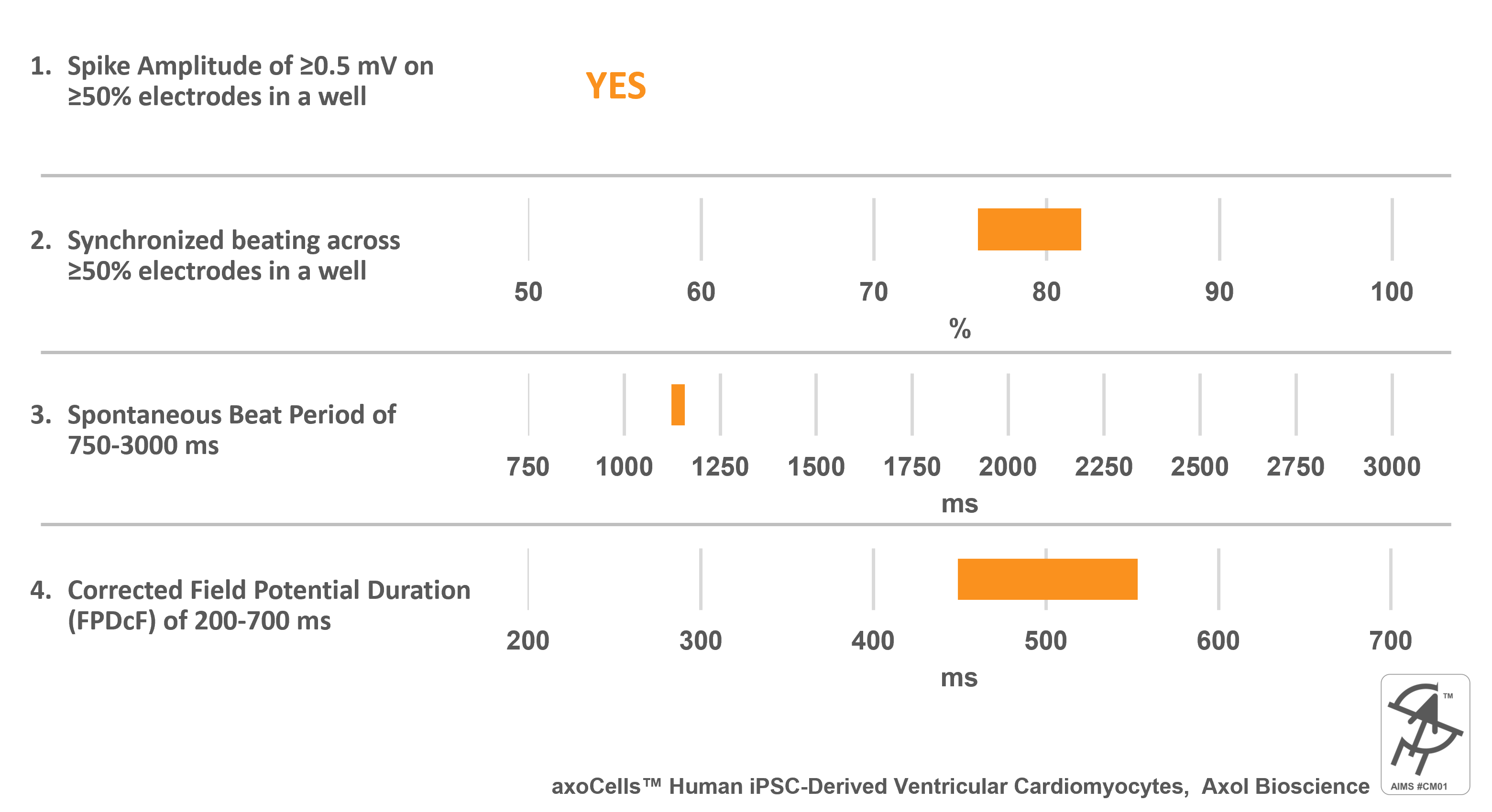
Cell specific information:
- Ventricular cardiomyocytes made from iPSCs generated from a 74 year-old male donor’s pulmonary fibroblasts.
- Validated against all 28 CiPA compounds
Recommended protocol:
Download Protocol
Recommend select publications:
- Kanade P P et al. Effects of low temperature on electrophysiology and mechanophysiology of human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs). Micro and Nano Syst Lett 9, 9 (2021). https://doi.org/10.1186/s40486-021-00135-2
Cardiosight®-S, NEXEL
Catalog Number(s): C-001 (2.5M cells), C-002 (5M cells)
AIMS CM#01 requirements:
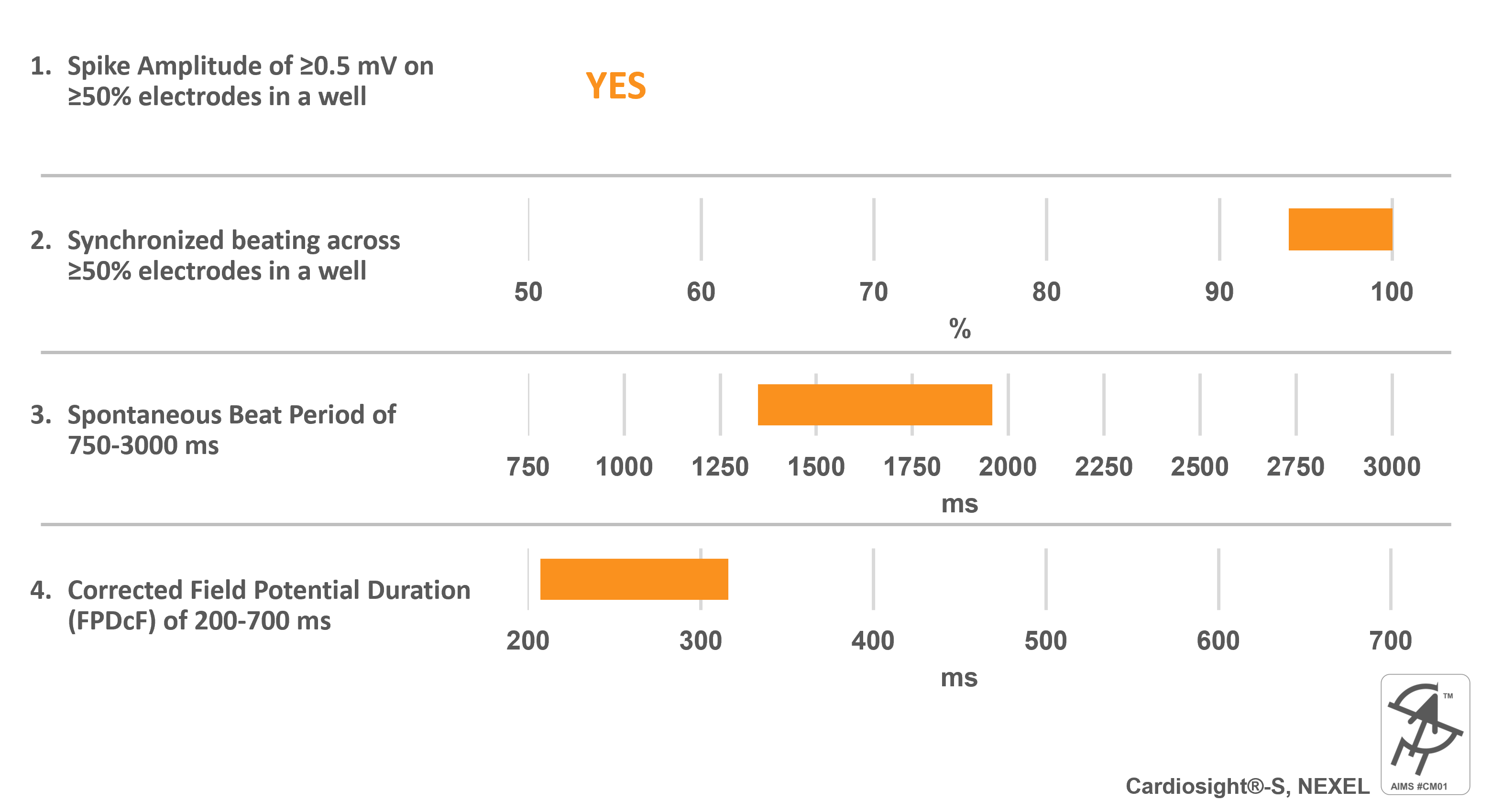
Cell specific information:
- Differentiated from normal foreskin fibroblasts (Caucacian)
- >95% pure Human Cardiomyocytes
- Mixture of Ventricular, Atrial, and Nodal cells (predominantly ventricular cells)
- Validated against 28 CiPA compounds
- Serum-free culture media
- Easy forming cardiac spheroids
Recommended protocol:
Download Protocol
- Maestro Pro MEA system is set to a temperature of 37 ºC with a CO2 concentration of 5.0%.
- Cells should be maintained in the antibiotic-free and serum-free, CMS-A media.
- It is recommended to perform MEA assay using Cardiosight®-S on Day 7 after thawing.
- On the day of MEA measurement, perform a full media change pre-warmed CMS-A media at least 3 hours prior to recording.
- Equilibrate MEA plate for 30 minutes prior to baseline recording on the Maestro Pro MEA system.
- All the batches of Cardiosight®-S are validated against negative control (DMSO 0.1%) and positive control (Dofetilide, E4031, Nifedipine, and Mexiletine). Detailed percent changes in FPDcF and Spike amplitude in response to these compounds are provided in the CoA.
Recommend select publications:
- Kim JS et. al. Impact of High-Dose Irradiation on Human iPSC-Derived Cardiomyocytes Using Multi-Electrode Arrays: Implications for the Antiarrhythmic Effects of Cardiac Radioablation Int J Mol Sci. 2021 Dec 29;23(1):351. doi: 10.3390/ijms23010351.
- Park SJ et. al. Discovery of Novel Sphingosine-1-Phosphate-1 Receptor Agonists for the Treatment of Multiple Sclerosis. J Med Chem. 2022 Feb 24;65(4):3539-3562. doi: 10.1021/acs.jmedchem.1c01979.
- Hwang M et. al. The three-dimensionality of the hiPSC-CM spheroid contributes to the variability of the field potential. Front Physiol. 2023 Mar 21:14:1123190. doi: 10.3389/fphys.2023.1123190.
CardioEasy® Human Cardiomyocytes, Cellapy
Catalog Number(s): CA2201106
AIMS CM#01 requirements:
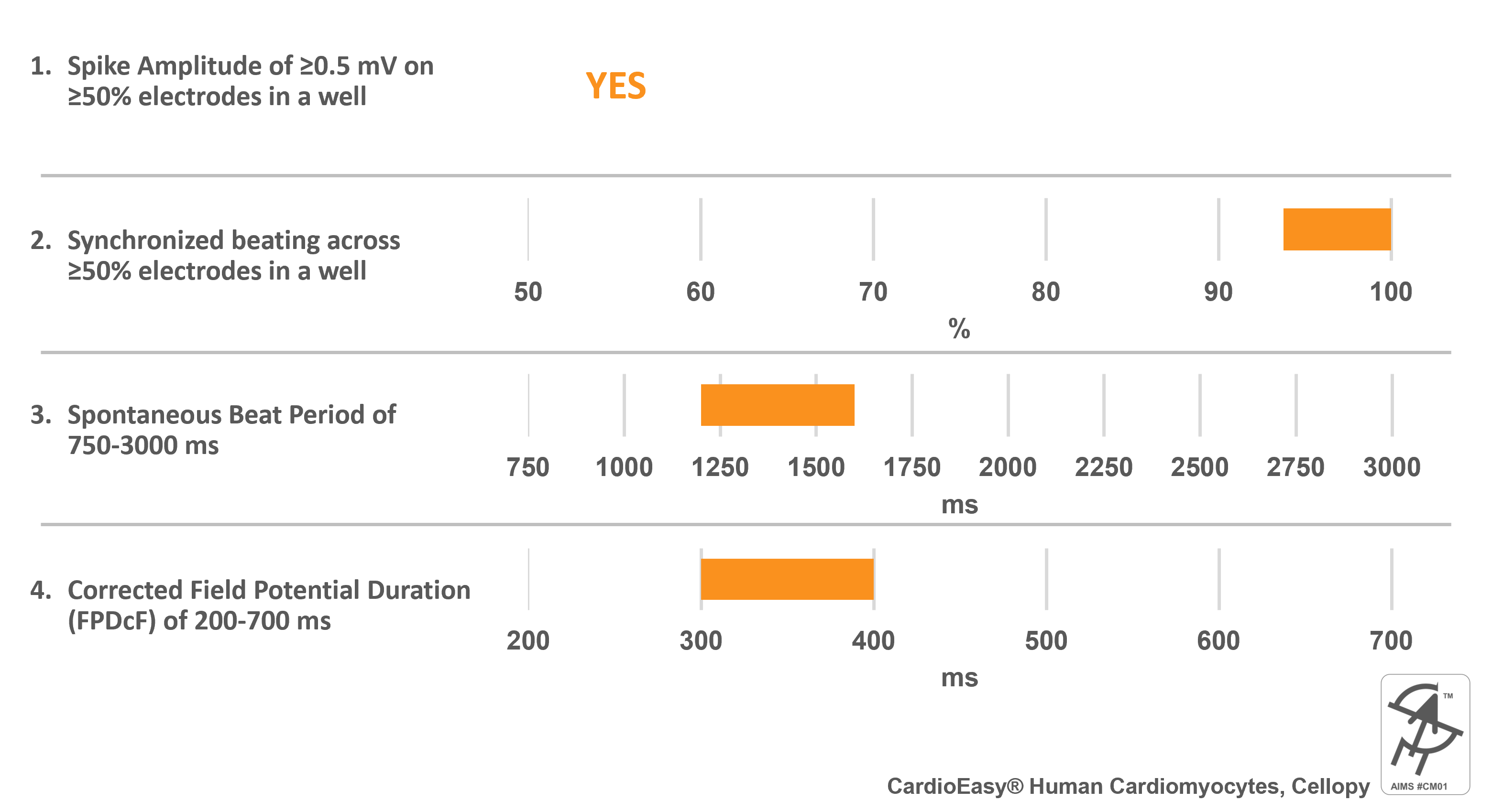
Cell specific information:
- Mixture of Ventricular, Atrial, and Nodal cells (predominantly ventricular cells)
- Chemically defined media used for culture and differentiation of hiPSCs
Recommended protocol:
Download Protocol
- Maestro MEA system is set to a temperature of 37 ºC with a CO2 concentration of 5.0%.
- Cells should be maintained in CardioEasy® Cardiomyocytes Maintenance Medium (CA2015002).
- A full-medium change overnight prior to baseline acquisition.
- Equilibrate MEA plate for 10 minutes prior to baseline recording on Maestro MEA system.
- On the day of MEA measurement, perform a full media change pre-warmed CMS-A media at least 3 hours prior to recording.
- Recommended experimental window for testing CardioEasy® Cardiomyocytes is Day 5-7 post-thaw.
- Recommended positive control compounds are Dofetilide (10 nM). hERG block prolongs FPDc by at least 20-30% after 15 min post-dose recording.
Recommend select publications:
- Li Y et. al. Frameshift variants in C10orf71 cause dilated cardiomyopathy in human, mouse, and organoid models. J Clin Invest. 2024; 134(12):e177172. doi: 10.1172/JCI177172.
- Li Y et. al. Zinc Oxide Nanoparticles Induce Mitochondrial Biogenesis Impairment and Cardiac Dysfunction in Human iPSC-Derived Cardiomyocytes. Int. J. Nanomed. 2020:15 2669–2683.
>> Reviewers:
Axion BioSystems would like to thank the following reviewers who helped shape this AIMS:
- Cristina Altrocchi - Principal Scientist Safety Pharmacology, J&J Innovative Medicine, Belgium
- Devon Guerrelli – Researcher, Children’s National Research Institute, USA
- Adam Hill - Head, Computational Cardiology Laboratory, Deputy Director Victor Chang Cardiovascular Innovation Centre, Australia
- Nikki Posnack - Principal Investigator, Children’s National Research Institute, USA
- Chris Strock - Vice President US ADMET Operations, Cyprotex, USA
- Takashi Yoshinaga - Executive Director, Advanced Biosignal Safety Assessment, Eisai, Japan
- Mike Clements - SVP, Axion BioSystems, USA
>> Acknowledgements:
Axion BioSystems would like to thank the following for their help reviewing this AIMS: Daniel Millard, Stacie Chvatal, Anthony Nicolini, Parker Ellingson, and Rika Yamazaki.