Benefits
>> Develop highly standardized sensory neuronal culture and analysis.
>> Screen acute or chronic peripheral nervous system (PNS) functional responses using compartmentalized high throughput electrophysiological recording.
>> Initialize innervation of organ using neurons as a bio-digital sensor.
>> Start predicting clinical outcomes based on reference compounds.
Overview
Neurons process their inputs through firing or not firing, which can be equated to producing zeros and ones. Since every single organ in the body is innervated, the peripheral nervous system (PNS) can essentially by seen as a distributed network of bio-digital sensors.
In this application note, we show how combining NETRI’s NeuroFluidics™ compartmentalized microelectrode array (MEA) platform with Axion’s BioSystems noninvasive Maestro MEA platform for hands-free electrophysiology has allowed us to develop a methodology for turning sensory neurons into reliable universal bio-digital sensors.
We show that by cultivating and fluidically isolating human induced pluripotent stem cell-derived (hiPSC) sensory neuron somas from their endings and stimulating the latter with reference compounds, we can repeatably record electrophysiological responses in a geometrical configuration that models human physiology. This can be used to screen novel compounds in wide ranging PNS applications ranging from dermato-cosmetics to pain management.
We finally show that multi-dimensional metric analysis can pave the way for compound fingerprinting through digital signatures and the emergence of digital libraries for advanced screening based on sought (or unsought) functional outcomes.
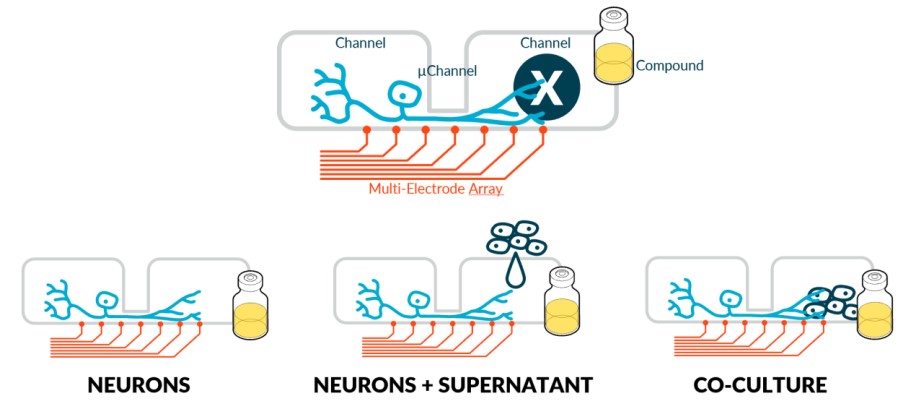
Results
Hardware (HW) & Software (SW) platform
We used the NeoBento™ standard SBS format outfitted with up to 16 DuaLink™ MEA chips, that combine NETRI’s microfluidic architectures with a bespoke MultiElectrode Array (MEA) surface from Axion BioSystems. Metrics are extracted using NETRI’s UpLink™ software piloting Axion BioSystems’ AxIS Navigator. Extracted data was processed in NETRI’s database DataLink. This HW/SW platform allows users to fluidically isolate somas from their endings (and eventually target organs) through microchannels. It can also perform compartmentalized functional activity recording, specifically in the ultra-sensitive µchannels that host the sensory neuron axons that ferry data from the sensed compartment to the sensing compartment.

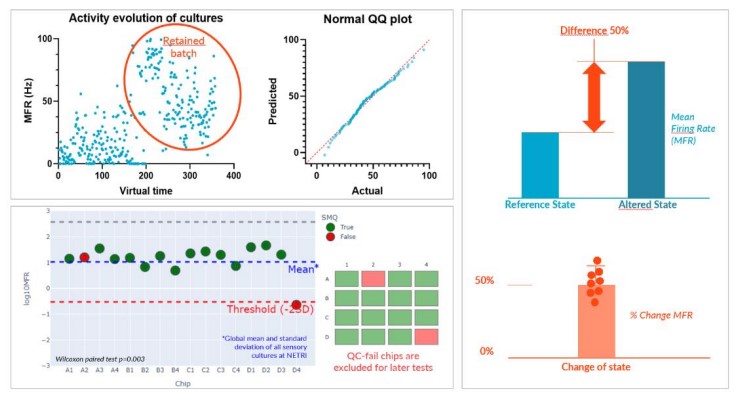
Sensor Repeatability
Achieving repeatability is critical to the development of a good sensor. First, we performed supplier batch selection and characterization with our partner Axol Bioscience and their axoCells™ Sensory Neurons.
Second, we adapted a single standard culture protocol for our DuaLink™ MEA chips to be used across all experimentations. Sensory neurons were seeded only in Channel 1 (Ch1) of the coated DuaLink™MEA (PDL + Surebond-XF) and cultured over 20 days.
Third, we developed a standardized quality control (QC) method applied to each culture based on a nominal basal MEA recording to exclude outliers (based on Negri et al. ENEURO.0080- 19.2019 paper).
Finally, MEA recording being non-destructive, our protocol measures the relative change within a single chip, between a reference state (basal activity) and an altered state (added compound). This self-referencing approach mitigates (inevitable) culture variability remaining after QC and allows statistical analysis.
Sensor Sensitivity
Sensor sensitivity is key to assess the ability of the sensor to react different stimuli, and to do so gradually without cut-off thresholds (within a range).
A – Time Response
The Time Response method, which is suitable when looking at a chronic, or at least long lasting, effect, records activity over two consecutive periods: reference state period and a subsequent altered state period where a compound was added. These can be spaced a few minutes or hours/days apart depending on the context of use.
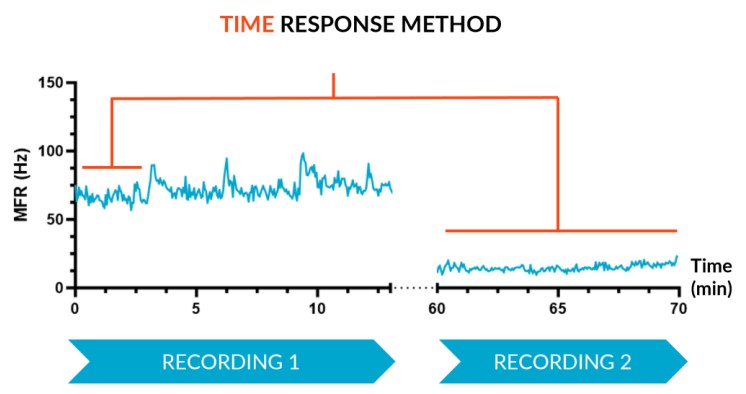
We subjected our cultures to standard stimuli in Channel 3 (Ch3) that span a range of applications, and showed a statistically significant response in each case, based on DuaLink™ MEA microchannel metrics alone.
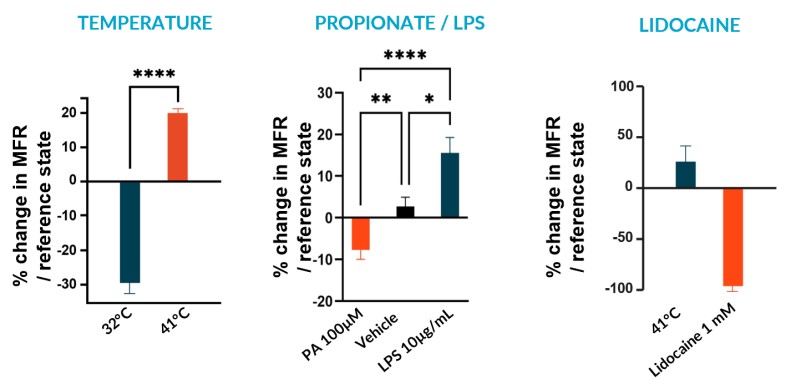
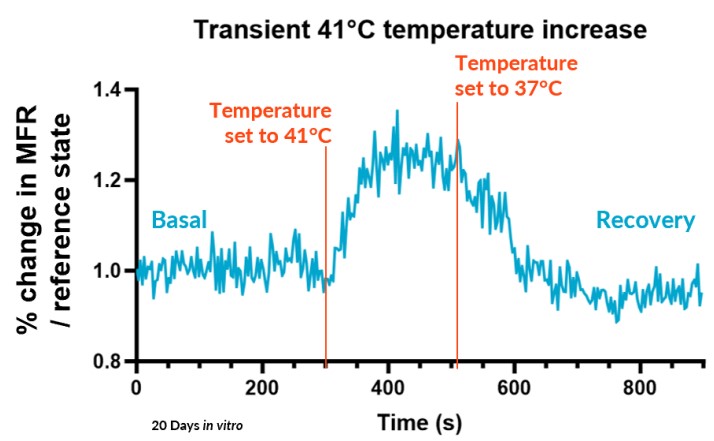
B – Fast Response
The Fast Response method, suitable for transient effects, records real-time activity for a single period for altered state (a compound) and reference states baselines (vehicle and positive control).
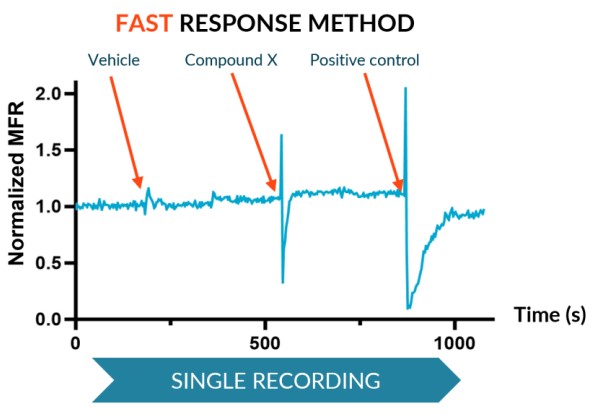
We subjected our cultures to standard stimuli in Channel 3 that span a range of applications, and showed a statistically significant response in each case, based on DuaLink™ microchannel metrics alone.
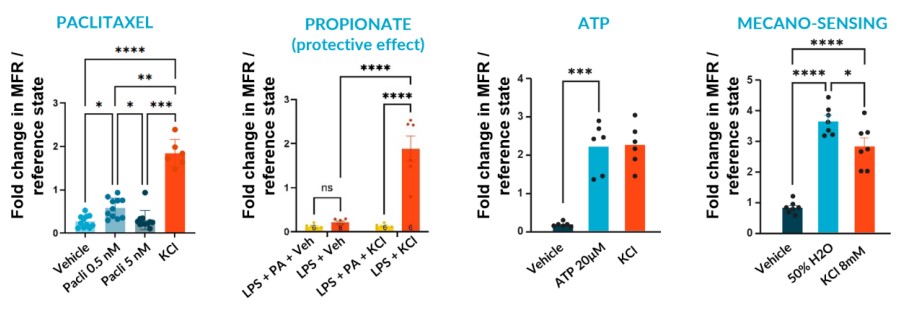
Sensor sensitivity can already serve multiple applications using application-specific altered state references. This is especially enabled for sensory neurons by recording activity in the microchannel that connects compartments.
Sensor Specificity
Sensitivity is limited in its ability to discriminate between altered states. We can therefore leverage the full gamut of metrics that MEA offers to distinguish seemingly alike signals from each other and confer specificity to our sensors. We conducted a preliminary test using two reference compounds, veratridine and lidocaine, and their corresponding vehicles, DMSO (Vd) and H2O (Vh), added to neurites in Ch3 only.
Out of all MEA metrics, we isolated the ones that have a Pearson correlation score less than 0.8, among which we select the ones that have the highest Information gain for a similarity matrix or ran a PCA for plotting. We also isolated the microchannels for being the source of the most discriminating metrics. Based on the selected metrics and electrode compartments, we managed to discern meaningful differences between all the states. Of note, this method does also discriminate between the two vehicles.
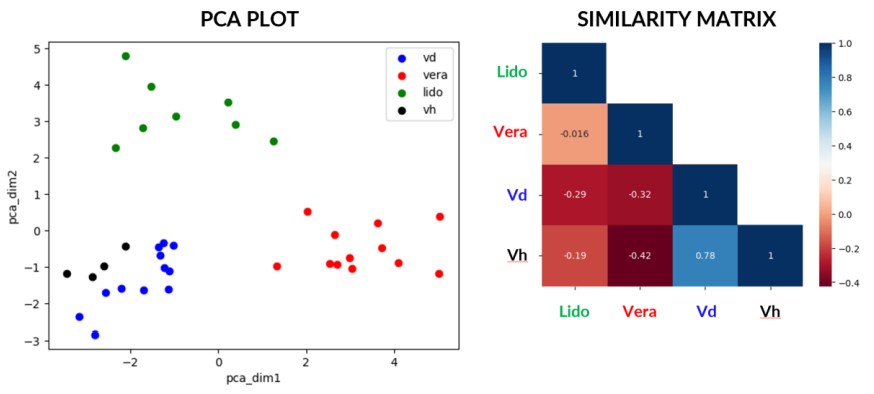
These early findings above show the ability to plot meaningful metrics that show discriminating patterns where simple mean firing rate readouts cannot. This suggests that we can achieve sensor specificity and pave the way for digital signatures of altered states.
Conclusion and Perspectives
The central notion emerging from this neuron-as-a-sensor approach is the power that well-calibrated, fluidically isolated neuronal cultures bring to the standardization of PNS electrophysiological functional analysis.
This opens the way to assessing the effects of a compound on a disease, a topical agent on the skin, a nutrient in the gut, or the repositioning of a drug, using neurons-as-bio-digital-sensors as an impactful and agnostic platform solution in a wide variety of applications that extend far beyond neuroscience. By defining digital signatures as an n-dimensional MEA-metric matrix of proportional ratios between a reference state (healthy) and a target state (altered) of multiple replicates, we will be able to drastically diminish the impact of an individual culture’s variability and show the quantification of drug candidate effects on a scale defined by reference compounds. More profoundly, this will be done non-destructively on acute or chronic timescales, without the use of imagery or chemical assays.
Resources
- Operating Protocol_axoCells™ Sensory Neurons.
- Application Note, Compartmentalized culture of primary or hiPSC-derived neurons using an MEA-capable high-throughput organs-on-chip platform, 2023.
- Poster, Compartmentalized MEA Pain(s)-on-chip platform, 2024.
- Poster, Digital Signature Library: using neurons as universal bio-digital sensors, 2024
